Motivation
All projects focus on developing prognostic tools for cancer or predictive tools for therapeutic efficacy and treatment emergent adverse events (TEAEs)
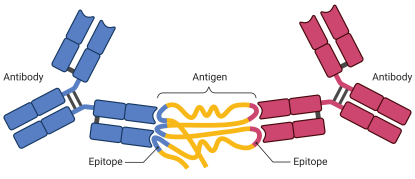
Protecting me from myself: Autoantibodies
An antibody is a protein that is made by the immune system in response to a substance called an antigen. The antigen is usually a threat such as a pathogen or toxin. The antibody binds directly to the antigen to help destroy the antigen. An autoantibody is an antibody made against substances formed by a person’s own body. Autoantibodies can directly destroy cells that have the antigens on them or can make it easier for other white blood cells to destroy them. Some autoimmune diseases are caused by autoantibodies. We are developing an autoantibody matrix platform that contains ≥ 20,000 potential human antigen targets that will allow us to screen biospecimens for the presence of autoantibodies. We are developing tools for statistical assessment and data visualization to (1) Identify autoantibodies and their associated biological pathways, (2) Identify autoantibodies that can be used for assay normalization and QA/QC assessment, (3) Quantitate autoantibody expression based on GST-tag expression and other autoantibody matrix platform controls, and (4) Determine the role of autoantibodies in health and disease. Multiple projects are available for interested students and advanced trainees.
Clones R Us: Deep dive into immunarch for immune repertoire profiling
The immune system attacks and destroys threats (e.g., toxins, pathogens, cancer) through pattern recognition of antigens (e.g., proteins) that comprise the threat. B cells and T cells respond to that pattern recognition by making clones of themselves that are specific for recognition of the antigen. When we treat patients with various therapies, those clones are either eliminated, expanded, or have no response to the treatments. Developing visual tools that can identify clones before treatment and after treatment with a display of the kinetics of their behavior will shed light on what treatments are doing to these immune populations when treatments work and when they do not work. Development of tools in programing languages such as R and Java could be a helpful for visualization and statistical assessment of the kinetics of the behaviors. Multiple projects are available for interested students and advanced trainees.
Essence of Ground Truth: Real-world scenarios
Histology, the study of microscopic structures in patient tissue, is a pathologist’s and clinician’s first step to diagnosing and monitoring many diseases. Patient tissues are stained with vibrant dyes, such as hematoxylin and eosin (H&E), that highlight components of the tissue such as cells, vessels, and architectural features. Artificial Intelligence Machine Learning tools are being developed to identify and quantitate components and features in digitally scanned images of H&E stained tissue. However, comparative ground truth or reference data that are required to produce effective recognition of real-world scenarios are lacking. Using a custom-built microscopy platform with and without augmented reality capabilities for reader studies, we are developing ground truth datasets, imaging software (acquisition, visualization, assessment), statistical packages, and educational training tools for H&E stained tissue assessment in collaboration with the U.S. Food and Drug Administration (FDA). Multiple projects are available for interested students and advanced trainees.
Location, location!!!!: Spatial Dynamics of TIME
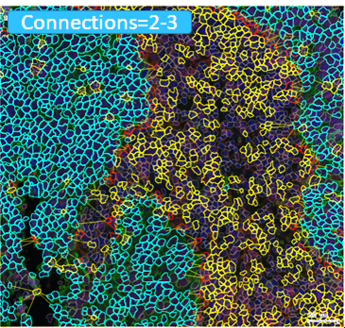
The right tumor immune microenvironment (TIME) is critical for elimination of cancer cells. Immunohistochemistry and immunofluorescence staining of histology tissue are used to assess multiple cellular (e.g., immune cells) and structural (e.g., extracellular matrix) targets. We have developed several staining methods and software tools to evaluate >25 targets (e.g., cell proteins) simultaneously in a single tissue section using different color chromogens and fluorochromes. The distribution of each target is assessed in the tissue through creation of unique spectral wavelength profiles for each chromogen or fluorochrome associated with the target of interest. These unique spectral wavelength profiles allow for linear unmixing of the targets of interest into separate channels which enables the quantitation of the targets on each cell and assessment of spatial relationships within a scanned image of the chromogen, fluorochrome, or standard H&E stained histology tissue. We are developing methods and models utilizing deep learning neural networks and unconventional software programs to identify and visualize the changes in spatial relationships over distances from key components within TIME. Multiple projects are available for interested students and advanced trainees.
Best of many worlds: Amalgamations - Proteogenomic profiling
TP53 is the gene that encodes p53, a protein that regulates cell division by keeping cells from growing and dividing too fast or uncontrollably. TP53 mutations are high in human papilloma virus (HPV) negative head and neck small cell carcinomas (HNSCCs). These mutations may be associated with specific changes in the tumor immune microenvironment. Phosphoproteomics, proteomics, transcriptomics, DNA methylation, somatic copy number alterations, and mutational analysis have been performed in a large HPV HNSCC cohort to address this question. Integration of data from multiple -omic platforms may help to provide insights that can lead to more precise approaches to treatment. We are generating spatial proteogenomics data from a real-world cohort of patients and creating software tools and methods to analyze and visualize the data to shed light on immune biomarkers and biological pathways associated with clinical outcome. Multiple projects are available for interested students and advanced trainees.
Best of many worlds: Amalgamations - N of 1 profiling
For decades we have relied on aggregate data from groups of patients to inform on prognostic and predictive biomarkers, diagnosis, and ultimately treatment decisions for an individual patient. Technology, computer science, computational biology, bioinformatics, and related fields have significantly improved to a point that we can now start to create methods, models, and tools to directly interrogate and make informed decisions on individual patient data. This will allow us to finally truly move towards personalized medicine in which each patient serves as their own control. We are integrating clinical data and data from multiple -omic platforms. We are creating models, software tools, and methods to extract, analyze, visualize, and interpret individual patient data for N of 1 profiling. Multiple projects are available for interested students and advanced trainees.
My voice: Patient reported outcomes (PROs)
Cancer patients are commonly treated with drug therapies that produce adverse reactions called treatment emergent adverse events (TEAEs). TEAEs can be mild to severe. There is a lack of good patient-friendly methods and tools to help patients to report their TEAEs. To address this gap, platforms are being developed to help capture types and severity of TEAEs directly from patients. Multiple projects are available for interested students and advanced trainees.
Digging for Buried Treasure: Data Mining
Electronic medical records (EMR), clinical data repositories, -omics repositories from clinical trials, and imaging repositories are excellent sources of clinical reported outcomes (ClinROs). Data is often a combination of structured and unstructured entries that require manual procedures and/or data mining algorithms to extract, organize, process, and analyze the data. The data output can help to address questions regarding TEAEs and be a source of new exciting hypothesis. Multiple projects are available for interested students and advanced trainees.
Rightsizing expectations: Renalase for Breast Cancer
Renalase is a protein that is secreted in the blood by the kidneys. Studies have shown that Renalase expression is significantly upregulated in multiple tumor microenvironments (pancreatic, bladder, melanoma, breast) and is inversely correlated with overall survival in these patients. There is evidence that it promotes cancer cell survival. The exact role of Renalase in general is unclear. However, it is known that it binds to the ATP2B4/PCMA4b receptor (a calcium ion transporter), engages the MAPK and PI3K pathways, and is regulated by STAT3. If Renalase is a survival factor for breast cancer, abnormal regulation of Renalase expression and signaling could provide a survival advantage for breast cancer cells, promote tumor growth, or promote breast cancer survival conditions in metastatic lesions or during dormancy. Any of these survival advantages could impact responses to therapy including immunotherapy. Better understanding of Renalase function could have therapeutic implications for its use for the management of breast cancer. Specifically, targeting Renalase could lower the threshold for sensitivity to drug therapies. Multiple projects are available for interested students and advanced trainees.